Neurological Disease Insights Through Translatomics
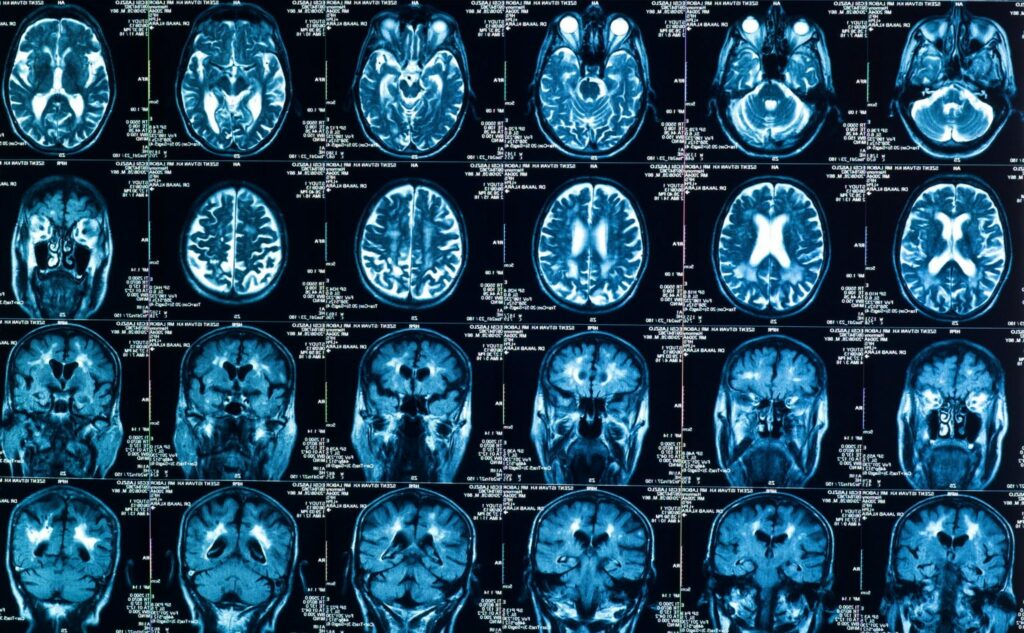
Translatomics in Action: Neurological Disease The dysregulation of translation in the central nervous system (CNS) has been linked to multiple neurodegenerative diseases, making translatomics a particularly important set of tools for learning and understanding more about neurological disease. Among the diseases in which translation has been implicated are amyotrophic lateral sclerosis (ALS) and frontotemporal lobar […]
Neurological Disease Insights Through Translatomics Read More »